Do Chromosomes Change Into Chromatin Again
Not only are the genomes of most eukaryotes much more complex than those of prokaryotes, but the DNA of eukaryotic cells is also organized differently from that of prokaryotic cells. The genomes of prokaryotes are contained in single chromosomes, which are usually circular DNA molecules. In contrast, the genomes of eukaryotes are composed of multiple chromosomes, each containing a linear molecule of DNA. Although the numbers and sizes of chromosomes vary considerably between different species (Table 4.2), their basic structure is the same in all eukaryotes. The DNA of eukaryotic cells is tightly bound to small basic proteins (histones) that package the DNA in an orderly way in the cell nucleus. This task is substantial, given the DNA content of most eukaryotes. For example, the total extended length of DNA in a human cell is nearly 2 m, but this DNA must fit into a nucleus with a diameter of only 5 to 10 μm. Although DNA packaging is also a problem in bacteria, the mechanism by which prokaryotic DNAs are packaged in the cell appears distinct from that of eukaryotes and is not well understood.
Table 4.2
Chromosome Numbers of Eukaryotic Cells.
Chromatin
The complexes between eukaryotic DNA and proteins are called chromatin, which typically contains about twice as much protein as DNA. The major proteins of chromatin are the histones—small proteins containing a high proportion of basic amino acids (arginine and lysine) that facilitate binding to the negatively charged DNA molecule. There are five major types of histones—called H1, H2A, H2B, H3, and H4—which are very similar among different species of eukaryotes (Table 4.3). The histones are extremely abundant proteins in eukaryotic cells; together, their mass is approximately equal to that of the cell's DNA. In addition, chromatin contains an approximately equal mass of a wide variety of nonhistone chromosomal proteins. There are more than a thousand different types of these proteins, which are involved in a range of activities, including DNA replication and gene expression. Histones are not found in eubacteria (e.g., E. coli), although the DNA of these bacteria is associated with other proteins that presumably function like histones to package the DNA within the bacterial cell. Archaebacteria, however, do contain histones that package their DNAs in structures similar to eukaryotic chromatin.
The basic structural unit of chromatin, the nucleosome, was described by Roger Kornberg in 1974 (Figure 4.8). Two types of experiments led to Kornberg's proposal of the nucleosome model. First, partial digestion of chromatin with micrococcal nuclease (an enzyme that degrades DNA) was found to yield DNA fragments approximately 200 base pairs long. In contrast, a similar digestion of naked DNA (not associated with proteins) yielded a continuous smear of randomly sized fragments. These results suggested that the binding of proteins to DNA in chromatin protects regions of the DNA from nuclease digestion, so that the enzyme can attack DNA only at sites separated by approximately 200 base pairs. Consistent with this notion, electron microscopy revealed that chromatin fibers have a beaded appearance, with the beads spaced at intervals of approximately 200 base pairs. Thus, both the nuclease digestion and the electron microscopic studies suggested that chromatin is composed of repeating 200-base-pair units, which were called nucleosomes.

Figure 4.8
The organization of chromatin in nucleosomes. (A) The DNA is wrapped around histones in nucleosome core particles and sealed by histone H1. Nonhistone proteins bind to the linker DNA between nucleosome core particles. (B) Gel electrophoresis of DNA fragments (more...)
More extensive digestion of chromatin with micrococcal nuclease was found to yield particles (called nucleosome core particles) that correspond to the beads visible by electron microscopy. Detailed analysis of these particles has shown that they contain 146 base pairs of DNA wrapped 1.65 times around a histone core consisting of two molecules each of H2A, H2B, H3, and H4 (the core histones) (Figure 4.9). One molecule of the fifth histone, H1, is bound to the DNA as it enters each nucleosome core particle. This forms a chromatin subunit known as a chromatosome, which consists of 166 base pairs of DNA wrapped around the histone core and held in place by H1 (a linker histone).
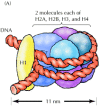
Figure 4.9
Structure of a chromatosome. (A) The nucleosome core particle consists of 146 base pairs of DNA wrapped 1.65 turns around a histone octamer consisting of two molecules each of H2A, H2B, H3, and H4. A chromatosome contains two full turns of DNA (166 base (more...)
The packaging of DNA with histones yields a chromatin fiber approximately 10 nm in diameter that is composed of chromatosomes separated by linker DNA segments averaging about 80 base pairs in length (Figure 4.10). In the electron microscope, this 10-nm fiber has the beaded appearance that suggested the nucleosome model. Packaging of DNA into such a 10-nm chromatin fiber shortens its length approximately sixfold. The chromatin can then be further condensed by coiling into 30-nm fibers, the structure of which still remains to be determined. Interactions between histone H1 molecules appear to play an important role in this stage of chromatin condensation.

Figure 4.10
Chromatin fibers. The packaging of DNA into nucleosomes yields a chromatin fiber approximately 10 nm in diameter. The chromatin is further condensed by coiling into a 30-nm fiber, containing about six nucleosomes per turn. (Photographs courtesy of Ada (more...)
The extent of chromatin condensation varies during the life cycle of the cell. In interphase (nondividing) cells, most of the chromatin (called euchromatin) is relatively decondensed and distributed throughout the nucleus (Figure 4.11). During this period of the cell cycle, genes are transcribed and the DNA is replicated in preparation for cell division. Most of the euchromatin in interphase nuclei appears to be in the form of 30-nm fibers, organized into large loops containing approximately 50 to 100 kb of DNA. About 10% of the euchromatin, containing the genes that are actively transcribed, is in a more decondensed state (the 10-nm conformation) that allows transcription. Chromatin structure is thus intimately linked to the control of gene expression in eukaryotes, as will be discussed in Chapter 6.

Figure 4.11
Interphase chromatin. Electron micrograph of an interphase nucleus. The euchromatin is distributed throughout the nucleus. The hetero-chromatin is indicated by arrowheads, and the nucleolus by an arrow. (Courtesy of Ada L. Olins and Donald E. Olins, Oak (more...)
In contrast to euchromatin, about 10% of interphase chromatin (called heterochromatin) is in a very highly condensed state that resembles the chromatin of cells undergoing mitosis. Heterochromatin is transcriptionally inactive and contains highly repeated DNA sequences, such as those present at centromeres and telomeres.
As cells enter mitosis, their chromosomes become highly condensed so that they can be distributed to daughter cells. The loops of 30-nm chromatin fibers are thought to fold upon themselves further to form the compact metaphase chromosomes of mitotic cells, in which the DNA has been condensed nearly 10,000-fold (Figure 4.12). Such condensed chromatin can no longer be used as a template for RNA synthesis, so transcription ceases during mitosis. Electron micrographs indicate that the DNA in metaphase chromosomes is organized into large loops attached to a protein scaffold (Figure 4.13), but we currently understand neither the detailed structure of this highly condensed chromatin nor the mechanism of chromatin condensation.

Figure 4.12
Chromatin condensation during mitosis. Scanning electron micrograph of metaphase chromosomes. Artificial color has been added. (Biophoto Associates/Photo Researchers Inc.)

Figure 4.13
Structure of metaphase chromosomes. An electron micrograph of DNA loops attached to the protein scaffold of metaphase chromosomes that have been depleted of histones. (From J. R. Paulson and U. K. Laemmli, 1977. Cell 12: 817.)
Metaphase chromosomes are so highly condensed that their morphology can be studied using the light microscope (Figure 4.14). Several staining techniques yield characteristic patterns of alternating light and dark chromosome bands, which result from the preferential binding of stains or fluorescent dyes to AT-rich versus GC-rich DNA sequences. These bands are specific for each chromosome and appear to represent distinct chromosome regions. Genes can be localized to specific chromosome bands by in situ hybridization, indicating that the packaging of DNA into metaphase chromosomes is a highly ordered and reproducible process.

Figure 4.14
Human metaphase chromosomes. (A) A light micrograph of human chromosomes spread from a metaphase cell. (B) Human chromosomes are arranged in pairs numbered from the largest (chromosome 1) to the smallest. The chromosomes shown are from a female, so there (more...)
Centromeres
The centromere is a specialized region of the chromosome that plays a critical role in ensuring the correct distribution of duplicated chromosomes to daughter cells during mitosis (Figure 4.15). The cellular DNA is replicated during interphase, resulting in the formation of two copies of each chromosome prior to the beginning of mitosis. As the cell enters mitosis, chromatin condensation leads to the formation of metaphase chromosomes consisting of two identical sister chromatids. These sister chromatids are held together at the centromere, which is seen as a constricted chromosomal region. As mitosis proceeds, microtubules of the mitotic spindle attach to the centromere, and the two sister chromatids separate and move to opposite poles of the spindle. At the end of mitosis, nuclear membranes re-form and the chromosomes decondense, resulting in the formation of daughter nuclei containing one copy of each parental chromosome.
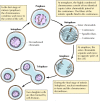
Figure 4.15
Chromosomes during mitosis. Since DNA replicates during interphase, the cell contains two identical duplicated copies of each chromosome prior to entering mitosis.
The centromeres thus serve both as the sites of association of sister chromatids and as the attachment sites for microtubules of the mitotic spindle. They consist of specific DNA sequences to which a number of centromere-associated proteins bind, forming a specialized structure called the kinetochore (Figure 4.16). The binding of microtubules to kinetochore proteins mediates the attachment of chromosomes to the mitotic spindle. Proteins associated with the kinetochore then act as "molecular motors" that drive the movement of chromosomes along the spindle fibers, segregating the chromosomes to daughter nuclei.
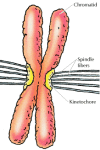
Figure 4.16
The centromere of a metaphase chromosome. The centromere is the region at which the two sister chromatids remain attached at metaphase. Specific proteins bind to centromeric DNA, forming the kinetochore, which is the site of spindle fiber attachment. (more...)
Centromeric DNA sequences have been defined best in yeasts, where their function can be assayed by following the segregation of plasmids at mitosis (Figure 4.17). Plasmids that contain functional centromeres segregate like chromosomes and are equally distributed to daughter cells following mitosis. In the absence of a functional centromere, however, the plasmid does not segregate properly, and many daughter cells fail to inherit plasmid DNA. Assays of this type have enabled determination of the sequences required for centromere function. Such experiments first showed that the centromere sequences of the well-studied yeast Saccharomyces cerevisiae are contained in approximately 125 base pairs consisting of three sequence elements: two short sequences of 8 and 25 base pairs separated by 78 to 86 base pairs of very AT-rich DNA (Figure 4.18A).

Figure 4.17
Assay of a centromere in yeast. Both plasmids shown contain a selectable marker (LEU2) and DNA sequences that serve as origins of replication in yeast (ARS, which stands for autonomously replicating sequence). However, plasmid I lacks a centromere and (more...)
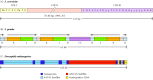
Figure 4.18
Centromeres of S. cerevisiae, S. pombe, and Drosophila melanogaster. (A) The S. cerevisiae centromere (CEN) sequences consist of two short conserved sequences (CDEI and CDEIII) separated by 78 to 86 base pairs (bp) of AT-rich DNA (CDEII). The sequences (more...)
The short centromere sequences defined in S. cerevisiae, however, do not appear to reflect the situation in other eukaryotes. More recent studies have defined the centromeres of the fission yeast Schizosaccharomyces pombe by a similar functional approach. Although S. cerevisiae and S. pombe are both yeasts, they appear to be as divergent from each other as either is from humans and are quite different in many aspects of their cell biology. These two yeast species thus provide complementary models for simple and easily studied eukaryotic cells. The centromeres of S. pombe span 40 to 100 kb of DNA; they are approximately a thousand times larger than those of S. cerevisiae. They consist of a central core of 4 to 7 kb of single-copy DNA flanked by repetitive sequences (Figure 4.18B). Not only the central core but also the flanking repeated sequences are required for centromere function, so the centromeres of S. pombe appear to be considerably more complex than those of S. cerevisiae.
Studies of a Drosophila chromosome have provided the first characterization of a centromere in higher eukaryotes (Figure 4.18C). The Drosophila centromere spans 420 kb, most of which (more than 85%) consists of two highly repeated satellite DNAs with the sequences AATAT and AAGAG. The remainder of the centromere consists of interspersed transposable elements, which are also found at other sites in the Drosophila genome, in addition to a nonrepetitive region of AT-rich DNA. Deletion of the satellite sequences and transposable elements, as well as the nonrepetitive DNA, reduced the activity of the centromere in functional assays. Thus, both repetitive and nonrepetitive sequences appear to contribute to kinetochore formation and centromere function.
Centromeres of humans and other mammals have not yet been defined by functional studies, but they have been identified by the binding of centromere-associated proteins. Mammalian centromeres are characterized by extensive regions of heterochromatin consisting of highly repetitive satellite DNA sequences. In humans and other primates the primary centromeric sequence is α satellite DNA, which is a 171-base-pair sequence arranged in tandem repeats spanning up to millions of base pairs. The α satellite DNA appears to play a role in centromere structure and function, since it has been found to bind centromere-associated proteins. However, the precise function of α satellite DNA, as well as the potential roles of other sequences in mammalian centromeres, remains to be established. Consistent with their large size, mammalian centromeres form large kinetochores that bind 30 to 40 microtubules, whereas only single microtubules bind to the centromeres of S. cerevisiae.
Telomeres
The sequences at the ends of eukaryotic chromosomes, called telomeres, play critical roles in chromosome replication and maintenance. Telomeres were initially recognized as distinct structures because broken chromosomes were highly unstable in eukaryotic cells, implying that specific sequences are required at normal chromosomal termini. This was subsequently demonstrated by experiments in which telomeres from the protozoan Tetrahymena were added to the ends of linear molecules of yeast plasmid DNA. The addition of these telomeric DNA sequences allowed these plasmids to replicate as linear chromosome-like molecules in yeasts, demonstrating directly that telomeres are required for the replication of linear DNA molecules.
The telomere DNA sequences of a variety of eukaryotes are similar, consisting of repeats of a simple-sequence DNA containing clusters of G residues on one strand (Table 4.4). For example, the sequence of telomere repeats in humans and other mammals is AGGGTT, and the telomere repeat in Tetrahymena is GGGGTT. These sequences are repeated hundreds or thousands of times, thus spanning up to several kilobases, and terminate with an overhang of single-stranded DNA. Recent results suggest that the repeated sequences of telomere DNA form loops at the ends of chromosomes, thereby protecting the chromosome termini from degradation (Figure 4.19).

Figure 4.19
Structure of a telomere. Telomere DNA loops back on itself to form a circular structure that protects the ends of chromosomes.
Telomeres play a critical role in replication of the ends of linear DNA molecules (see Chapter 5). DNA polymerase is able to extend a growing DNA chain but cannot initiate synthesis of a new chain at the terminus of a linear DNA molecule. Consequently, the ends of linear chromosomes cannot be replicated by the normal action of DNA polymerase. This problem has been solved by the evolution of a special mechanism, involving reverse transcriptase activity, to replicate telomeric DNA sequences. Maintenance of telomeres appears to be an important factor in determining the lifespan and reproductive capacity of cells, so studies of telomeres and telomerase have the promise of providing new insights into conditions such as aging and cancer.
Source: https://www.ncbi.nlm.nih.gov/books/NBK9863/
0 Response to "Do Chromosomes Change Into Chromatin Again"
Post a Comment